Ab Initio Protein Structure Reconstruction
Imagine trying to reconstruct eight different 3D objects when you only have thousands of unlabeled 2D shadows, you don’t know the viewing angles, and everything is buried in noise. This is the challenge structural biologists face when studying proteins with cryo-electron microscopy. Here’s how one algorithm solves this problem automatically, without any prior knowledge.
The Problem: Unmixing Molecular Shapes
Biological molecules are dynamic machines that constantly change shape (or “conformation”) to perform their functions. When scientists use cryo-electron microscopy (cryo-EM), a technique that images flash-frozen molecules with electron beams, they capture thousands of 2D projection images of these molecules from different angles. The core challenge is that a single sample contains a mix of these different conformations, a problem known as structural heterogeneity.
Traditional reconstruction algorithms often assume all projections come from one identical structure. This forces them to average dissimilar shapes, resulting in blurry reconstructions that obscure crucial details. Existing methods to separate conformations often require known reference structures or tedious manual classification, which is slow, biased, and can fail to identify new, undiscovered shapes.
Why This Matters: Proteins change shape to perform biological functions, such as enzyme catalysis, immune response, and viral infection. Understanding these conformational changes is crucial for drug design and disease treatment. Without accurate multi-conformation reconstruction, researchers might miss the exact protein state where a drug can bind effectively.
Our Solution: A Fully Automated Pipeline
We developed an automated algorithm that solves the ab initio reconstruction problem for heterogeneous samples. Here’s how it works:
-
Robust Clustering: We first use a variant of hierarchical clustering (single-linkage clustering) to group projections. Unlike standard k-means, this method creates “pure” clusters that contain projections from only one conformation, even in high-noise conditions.
-
Projection Denoising: The projections in each pure cluster are averaged and then cleaned using a patch-based PCA denoising algorithm. This produces a small set of clean, representative projections for each conformation.
-
Classification & Angle Estimation: We use a novel combination of techniques to sort the representative projections. A graph Laplacian-based algorithm reveals the underlying structure of the data, enabling us to infer the number of conformations present. We then use the Helgason-Ludwig consistency conditions to find initial estimates for the viewing angle of each projection.
-
Iterative Refinement: Finally, we alternately refine the viewing angles and reconstruct the structure for each class using filtered back-projection. This iterative process minimizes the reconstruction error, converging on a high-fidelity model of each distinct conformation.
Key Innovation
Our method’s strength comes from uniquely combining three mathematical frameworks:
- Graph Laplacian techniques to discover the number of distinct conformations automatically.
- Moment-based constraints (HLCC) to connect 2D projections to 3D orientations without prior knowledge.
- A robust statistical framework that withstands significant noise and correctly separates projections from different structures.
Crucially, the algorithm requires no prior structural information, templates, or even knowledge of how many conformations exist. In experiments on protein complexes like Lipase, our method successfully separated and reconstructed up to eight different conformations from a single dataset, even with high levels of noise (noise standard deviation up to 30% of the average signal value, i.e., σ=0.3a).
This approach enables researchers to discover novel protein states and understand molecular dynamics, free from the bias of existing structural knowledge.
Experimental Results
We validated our approach on simulated 2D datasets of protein complexes, demonstrating the algorithmic principles that extend to 3D reconstruction.
Noise Robustness
We tested our method on holo-GAPDHase (holo-glyceraldehyde-3-phosphate dehydrogenase), reconstructing three distinct conformations under progressively increasing noise levels. The results demonstrate remarkable noise tolerance: even at σ=0.3a (30% noise relative to the average signal), all three conformations are successfully recovered with reconstruction errors remaining under 32%.
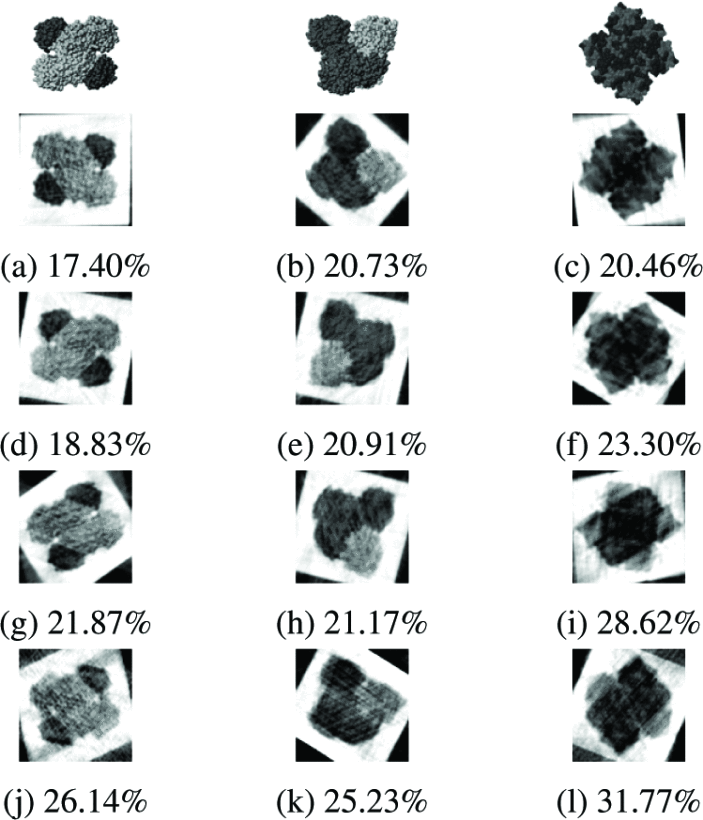
Three original conformations of holo-GAPDHase (top row) and their reconstructions at noise levels σ = 0.0a, 0.1a, 0.2a, and 0.3a (rows 2-5). RMSE values range from 17.40-20.73% (no noise) to 25.23-31.77% (high noise), demonstrating graceful degradation.
Multi-Conformation Recovery
In a challenging mixed-heterogeneity experiment, we combined projections from five Lipase conformations and three holo-GAPDHase conformations into a single unlabeled dataset. Using 80,000 noisy projections (σ=0.3a) with no prior classification information, the algorithm automatically identified all eight distinct structures and reconstructed each with consistent quality (RMSE: 16.81-25.15%). This validates the method’s ability to handle real-world scenarios where multiple unknown conformations coexist.
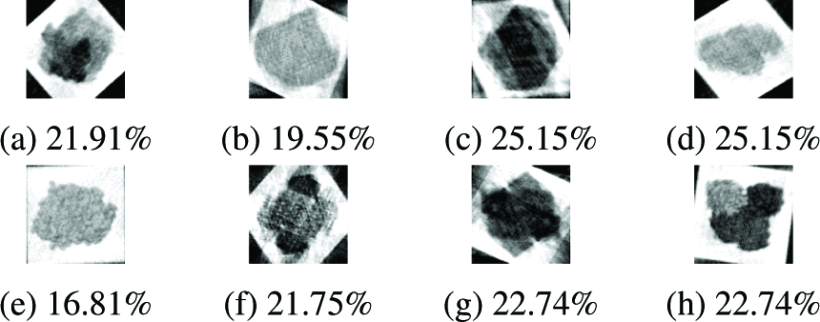
Reconstruction of eight structures automatically separated from a mixed dataset (5 Lipase + 3 holo-GAPDHase). All reconstructions achieved RMSE < 26% despite 30% noise and no prior knowledge of conformation count or structural templates.
The Bottom Line
This automated approach transforms cryo-EM heterogeneity analysis from a manual, reference-dependent process into a fully data-driven workflow. By automatically discovering and reconstructing up to eight distinct protein conformations from unlabeled, noisy data, the method enables researchers to study molecular dynamics without prior structural assumptions, opening doors to discovering novel therapeutic targets.
Paper Details
- Title: Ab Initio Tomography With Object Heterogeneity and Unknown Viewing Parameters
- Conference: IEEE International Conference on Image Processing (ICIP) 2019
- Presentation: Oral
- Authors: Arunabh Ghosh¹, Ritwick Chaudhry², Ajit Rajwade³
- Affiliations: ¹Dept. of EE, IIT Bombay; ²Adobe Research; ³Dept. of CSE, IIT Bombay
- Pages: 1257-1261
- DOI: 10.1109/ICIP.2019.8803750